Intro to Numpy and Simple Plotting¶
In this lesson you will be introduced to Numpy, and some simple plotting using pylab. A simple script will be introduced that opens a spreadsheet of data, computes some simple statistics, creates some simple plots.
Numpy = Numerical + Python¶
Numpy is the main building block for doing scientific computing with python. A wealth of information can be found here:
Numpy for Matlab users¶
As an object-oriented language, python may seem very different than matlab, and in many ways it is. However, for people with a Matlab background it can be fairly easy to switch to python using Numpy. These pages can be very useful to understand:
- which numpy function maps to a given matlab function
- difference between numpy array and numpy matrix
- how to index arrays, and fancy indexing
Script Deconstruction : numpy_example.py¶
In this example, I will open a file (this one uses colon : as a delimiter), and I will read in different fields so I can analyze and viualize the data.
import
import numpy as np
This is the main tool used in the rest of this script.
Parse the Header¶
I know my data file has a header row as its first row of data. So I plan to read this in and use this to help load in the rest of my data.
The beginning of the first row looks like this:
"BAC#":"Gender":"Handedness":"Years Ed": ...
As you can see
- Fields are separated by colon :
- There are extra quotes ” in the fields
So I will
- read in the first line
- strip unnecessary quotes
- split on the colon to create a list
- cast result to a numpy array
code:
infile = 'Cindee_11_9_10_v2.csv'
hdrstr = open(infile).readline()
hdrs = hdrstr.replace('"','').split(':')
hdrs = np.array(hdrs)
So Now I want to find the indicies of a field, lets say all the fields that have ‘BAC’ in them:
bac_ind = [np.where(hdrs==x) for x in hdrs if 'BAC' in x]
bac_ind = [x[0][0] for x in bac_ind] #simplify
bac_hdr_str = hdrs[bac_ind]
np.where returns indicies where a logical test on an array returns true
so this lets me find the indicies of the fields that have the term “BAC” in them....
Using loadtxt¶
Now that I have figured out what fields I want, Ill use np.loadtxt to load them into an array so I can work with the data:
np.loadtxt(fname, dtype=<type 'float'>, delimiter=None, skiprows=0, usecols=None)
np.loadtxt
- fname = filename of text file to open
- dtype = base datatype to cast data to
- delimiter = character used to specify boundry between fields
- skiprows = number of leading rows to skip (in case you need to skip headers)
- usecols = tuple indicating the specific columns of data you wish to extract
example:
mydata = (0,1,3,6,10,36)
dat = np.loadtxt(infile, delimiter=':',
dtype=np.str, usecols=mydata,
skiprows = 1)
array([['"BAC001"', '"F"', '18', '63', '30', '13'],
['"BAC002"', '"M"', '18', '70', '30', '7'],
['"BAC003"', '"F"', '14', '61', '30', '12'], ...
Im skipping the first row since I already have dealt with getting my headers. And Im casting the data to a dtype of string np.str because the columns I am grabbing have mixed types, I will re-cast them to the correct types later. I could also load different types separately to avoid this issue.
Grab MMSE out of the dat array, replace missing fields, and cast to float:
empty = np.where(dat[:,keys['mmse']]=='')
dat[empty,keys['mmse']] = np.nan
mmse = dat[:,keys['mmse']].astype(np.float)
array([ 30., 30., 30., 28., 30., 29., 28., 30., 28., 29., 29.,
28., 30., 30., NaN, 29., 29., 30., 30., 30., 30., 29.,
Simple Statistics¶
I may want to calculate a simple mean. But this will be an issue as I have NAN in the array. however, it is simple to mask the array:
mmse_mean = mmse[mmse>0].mean()
28.991735537190081
mmse[mmse>0].std()
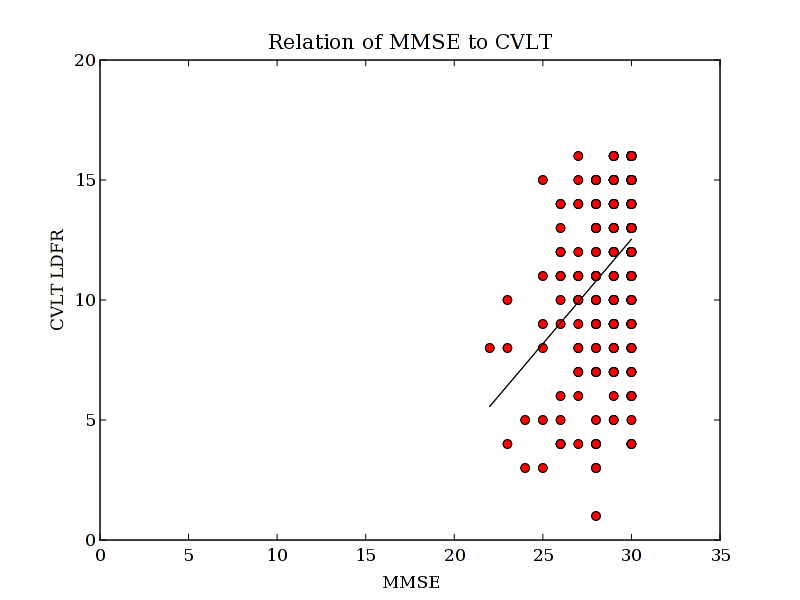
To make this more interesting...... I now get arrays of data for CVLT and age. For each of these, there may be different regions where subjects have missing fields. So if I want to compare two variables, I need to find the places where they overlap:
overlap2 = np.logical_and(mmse>0, cvlt>0)
I can now use some other numpy toos to see how two measures relate to each other.
polyfit:
p = np.polyfit(mmse[overlap2], cvlt[overlap2],1)
array([ 0.87076373, -13.59299814])
x = [mmse[overlap2].min(), mmse[overlap2].max()]
y = np.polyval(p, x)
least squares:
m = np.ones((mmse[overlap2].shape[0],2))
m[:,0] = mmse[overlap2]
results = np.linalg.lstsq(m, cvlt[overlap2])
print results[0]
array([ 0.87076373, -13.59299814])
Simple Plotting¶
In simple situations like this I use the -pylab directive of Ipython if I want to generate some simple plots:
ipython -pylab
It requires matplotlib to be installed, but is a powerful tool. There is a wonderful set of example plots to be found here:
http://matplotlib.sourceforge.net/gallery.html
So if we want to see our mmse, and cvlt data:
plot(mmse, cvlt, 'ro')
plot(x,y, 'k-')
axis([0,35,0,20])
title('Relation of MMSE to CVLT')
xlabel('MMSE')
ylabel('CVLT LDFR')
Homework¶
- Open a text file of (your own) data
- Parse the header
- Open the data and grab a subset of fields
- generate a simple statistic and create a simple plot
- save a png image
Email me if you get stuck, you have a few weeks
